Abstract
Background
B-cell precursor acute lymphoblastic leukemia (B-ALL) is a genetically heterogeneous group of acute leukemia with stage-specific phenotypes and cytogenetic features. Although the research on the molecular profile of B-ALL benefits diagnosis and risk stratification, the idiographic leukemogenesis beyond the transcriptome remains unknown. Genomic lesions in B-ALL frequently involve genes belonging to transcription factors, such as TCF3, EBF1, PAX5, and IKZF1. The investigation of dysregulated transcriptional networks behind various B-ALL subtypes may help unravel the specific process of leukemogenesis.
Methods
A random forest model was trained on a well-defined molecular subtype B-ALL cohort (n = 504) to improve the molecular classification. The subtype-specific transcriptional network was constructed by weighted correlation network analysis (WGCNA) once the B-ALL subtypes were genetically determined by the random forest model. Additionally, alternative splicing analysis from RNA-seq was emphasized since aberrant splicing events could lead to abnormalities in transcription factors or tumor suppressor genes.
Results
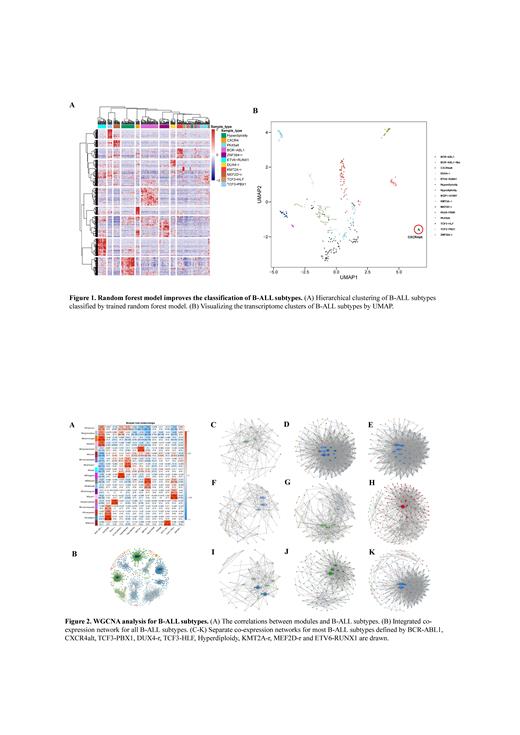
The random forest model performs well for the classification of most B-ALL subtypes (Figure 1A). It also benefits the classification of Ph-like B-ALL, which displays a gene expression profile similar to BCR-ABL1 B-ALL, as it achieves 100% accuracy on well-known Ph-like cases characterized by ABL-class gene fusions, PAX5-JAK2, EBF1-PDGFRB, and IGH-EPOR.
We successfully separated a candidate molecular subtype characterized by CXCR4 alteration (CXCR4alt) for the first time, through our novel classification model (Figure 1B). This newly identified CXCR4alt subtype accounts for 2% of B-ALL cases (11/504), characterized by CXCR4 C-terminal mutation R334X or FLNA overexpression. Both C-terminal mutation and upregulated FLNA contribute to delayed CXCR4 receptor internalization, enhanced CXCL12-CXCR4 signaling, and then continuously activates the downstream MAPK pathway. It is further supported by the high expression of the two oncogenic MAPK signaling pathway genes KIAA1549 and KIAA1549L from the co-expression network of CXCR4alt in these cases.
Transcriptional co-expression networks constructed by WGCNA and network hub genes for most B-ALL subtypes also help to elucidate the mechanism of leukemogenesis (Figure 2). We identified an alternative first exon of BLNK (BLNKaf) that leads to loss of function as a shared event in specific subtypes, such as BCR-ABL1, BCR-ABL1-like, and PAX5alt; while in pre-BCR signaling positive subtypes, such as TCF3-PBX1 and MEF2D-r, only express normal BLNK transcripts.
Discussion
By comprehensive transcriptome-based classification model and co-expression networks analysis, we identified a novel defined CXCR4alt subtype with an incidence of 2% in B-ALL. We also observed that BLNKaf might supply a practical marker for monitoring pre-BCR signaling. Our report emphasizes the role of transcriptome-based machine learning and WGCNA in mining the molecular mechanism of B-ALL. The molecular pathogenesis and clinical significance of these newly identified molecular subtypes and molecular abnormalities are worthy of further investigation.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal